SEOUL, November 10 (AJP) - KAIST researchers, working with Johns Hopkins University, have discovered how cells determine their direction of movement without any external signals, a finding that reveals the internal logic of cancer metastasis, immune responses, and tissue development.
The study, led by Professor Won Do Heo from the KAIST Department of Biological Sciences and Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering, together with Professor Gabsang Lee of Johns Hopkins University, identified an intrinsic "self-driving mechanism" that enables cells to decide which way to move. The results were published in Nature Communications on October 31.
To achieve this, the team developed a new imaging method called INSPECT (INtracellular Separation of Protein Engineered Condensation Technique) — a system that visualizes protein interactions inside living cells. Using INSPECT, they uncovered how specific combinations of signaling proteins dictate whether a cell moves straight ahead or turns in a new direction.
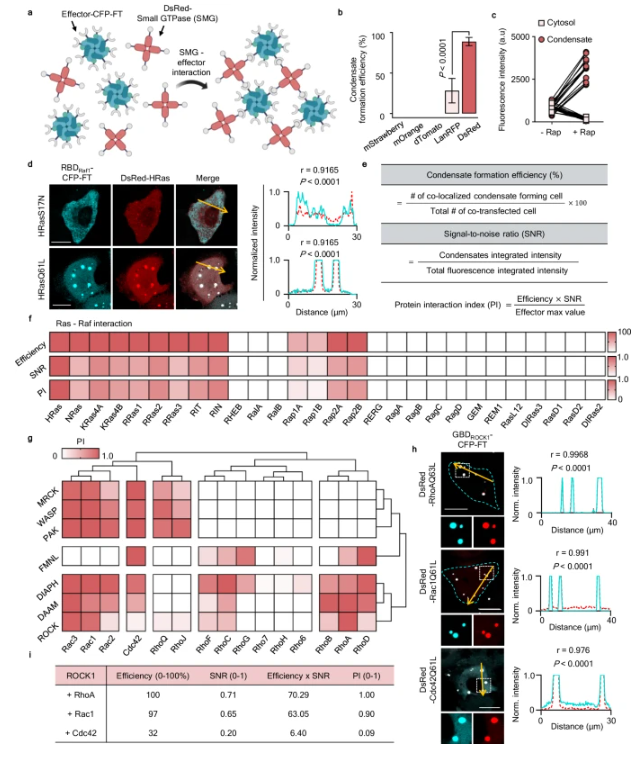
Their work focused on Rho GTPases — the Rac1, Cdc42, and RhoA proteins that control cell polarity and motility. The researchers found that these proteins do far more than divide a cell into front and rear sections. Instead, they form dynamic partnerships that act like a built-in navigation system. For instance, the Cdc42–FMNL complex drives linear motion, while the Rac1–ROCK pairing governs turning behavior.
INSPECT recreates a process known as phase separation, allowing scientists to see how proteins cluster and interact. By tagging proteins with ferritin and fluorescent DsRed, the team visualized condensate droplets forming as proteins bound to one another. Among 285 tested pairs, 139 were confirmed to interact in living cells.
When the team slightly altered one amino acid in Rac1 — preventing it from binding properly to its "steering handle" protein ROCK — cells lost their ability to turn and moved only in straight lines. Normal cells, by contrast, formed curved "arc stress fibers" that allowed them to pivot sharply when changing direction.
Additional experiments showed that while normal cells adjusted their speed in response to environmental cues, the modified Rac1F37W cells moved at a constant rate regardless of surroundings. This demonstrated that the Rac–ROCK axis finely tunes how cells sense and adapt to external environments.
Professor Heo said, "Our findings show that cell movement is not random but governed by an intrinsic program coordinated by Rho signaling proteins and their partners. INSPECT provides a powerful new tool for visualizing protein interactions in real time and will accelerate research on cancer metastasis, neural migration, and immune cell movement."
The paper's authors include Heeyoung Lee, Sangkyu Lee, Yeji Seo, Dongsan Kim, Yohan Oh, Juae Jin, Bobae Hyeon, Younghyun Han, Hyunjun Kim, Yong Jin Lee, Ho Min Kim, Gabsang Lee, Kwang-Hyun Cho, and Won Do Heo.
Copyright ⓒ Aju Press All rights reserved.